Resources
CHOIR
CHOIR (clustering hierachy optimization by iterative random forests) is a clustering algorithm for single-cell data. CHOIR applies a framework of permutation tests and random forest classifiers across a hierarchical clustering tree to statistically identify clusters that represent distinct populations. CHOIR is currently in beta. You can find everything you need on our website, in the preprint, or on our GitHub Repository.
ArchR
ArchR is a full-featured R package for processing and analyzing single-cell ATAC-seq data. ArchR provides the most extensive suite of scATAC-seq analysis tools of any software available. Additionally, ArchR excels in both speed and resource usage, making it possible to analyze 1 million cells in 8 hours on a MacBook Pro laptop. You can find everything you need on our website or in the publication.
Omni-ATAC
Omni-ATAC is a carefully optimized ATAC-seq method that provides improvements on data quality, cost, and generalizability. It works on every cell type that we have tested, from cell lines to flash frozen tissues. The original protocol can be found as Supplementary Protocol 1 of Corces et al. Nature Methods 2017 and an updated protocol for nuclei isolation from frozen tissue can be found on protocols.io.
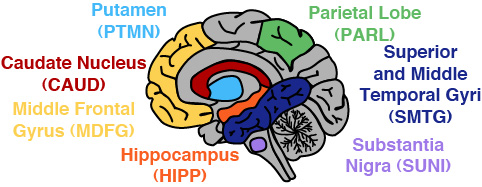
Human Brain scATAC-seq
Data from our 2020 publication on human brain scATAC-seq, HiChIP, and bulk ATAC-seq is publicly available through GEO. This data comes from cognitively healthy individuals. We have also set up a WashU Epigenome Browser track hub of cell type-specific chromatin accessibility as well as a HiGlass track hub of all bulk ATAC-seq datasets from the publication.
TCGA ATAC-seq
Data in the form of aligned hg38 BAM files from our 2018 collaboration with The Cancer Genome Atlas are available through the Genomic Data Commons Data Portal with approved access requested through dbGaP. Select processed files including bigwigs, counts matrices, and cancer type-specific peak calls are available through the TCGA publication page.